PITPNA Rabbit Polyclonal Antibody (C-term)
产品基本信息
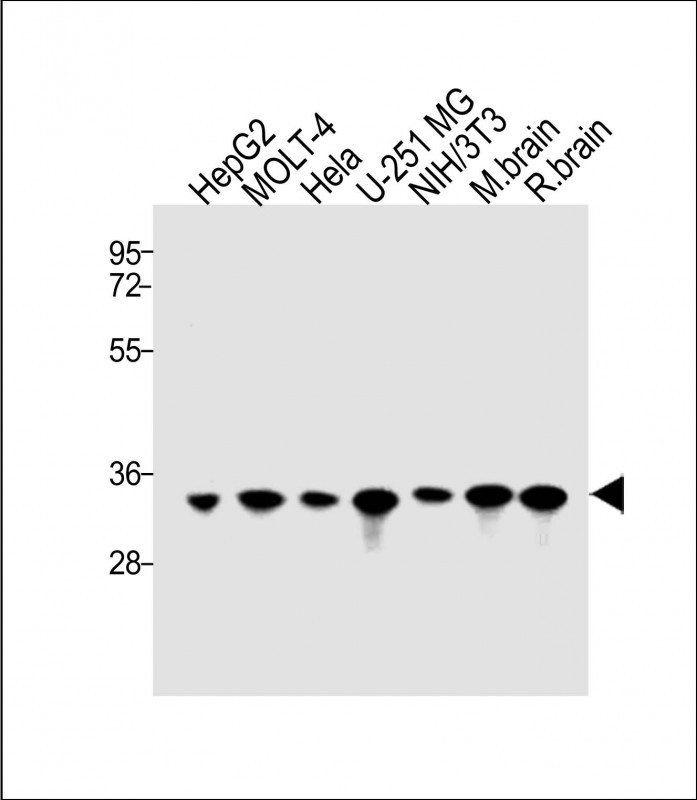
All lanes : Anti-PITPNA Antibody (C-term) at 1:1000 dilution Lane 1: HepG2 whole cell lysate Lane 2: MOLT-4 whole cell lysate Lane 3: Hela whole cell lysate Lane 4: U-251MG whole cell lysate Lane 5: NIH/3T3 whole cell lysate Lane 6: Mouse brain lysate Lane 7: Rat brain lysate Lysates/proteins at 15 μg per lane. Secondary Goat Anti-Rabbit IgG, (H+L), Peroxidase conjugated at 1/10000 dilution. Predicted band size : 32 kDa Blocking/Dilution buffer: 5% NFDM/TBST.
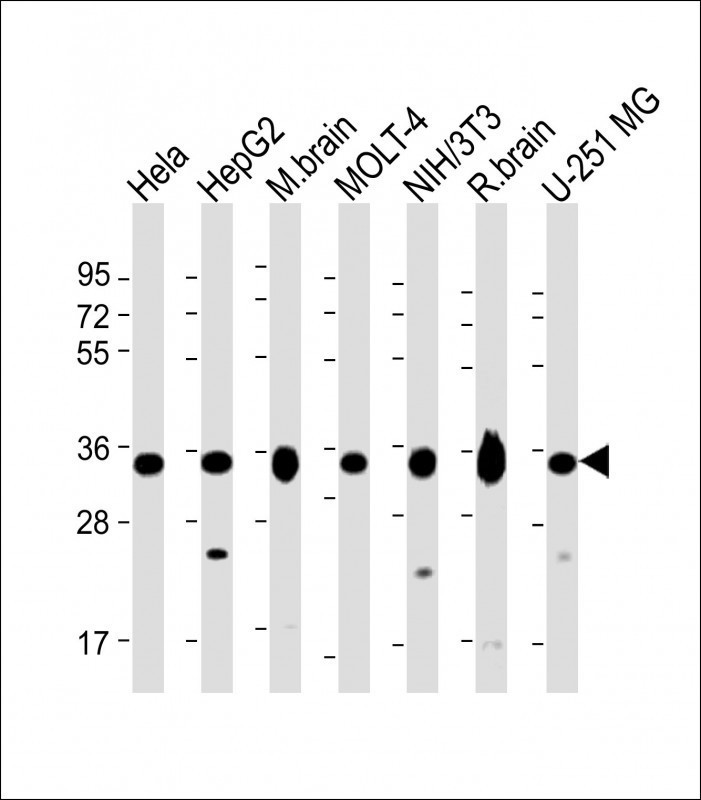
All lanes : Anti-PITPNA Antibody (C-term) at 1:2000 dilution Lane 1: Hela whole cell lysate Lane 2: HepG2 whole cell lysate Lane 3: mouse brain lysate Lane 4: MOLT-4 whole cell lysate Lane 5: NIH/3T3 whole cell lysate Lane 6: rat brain lysate Lane 7: U-251 MG whole cell lysate Lysates/proteins at 20 μg per lane. Secondary Goat Anti-Rabbit IgG, (H+L), Peroxidase conjugated at 1/10000 dilution. Predicted band size : 32 kDa Blocking/Dilution buffer: 5% NFDM/TBST.
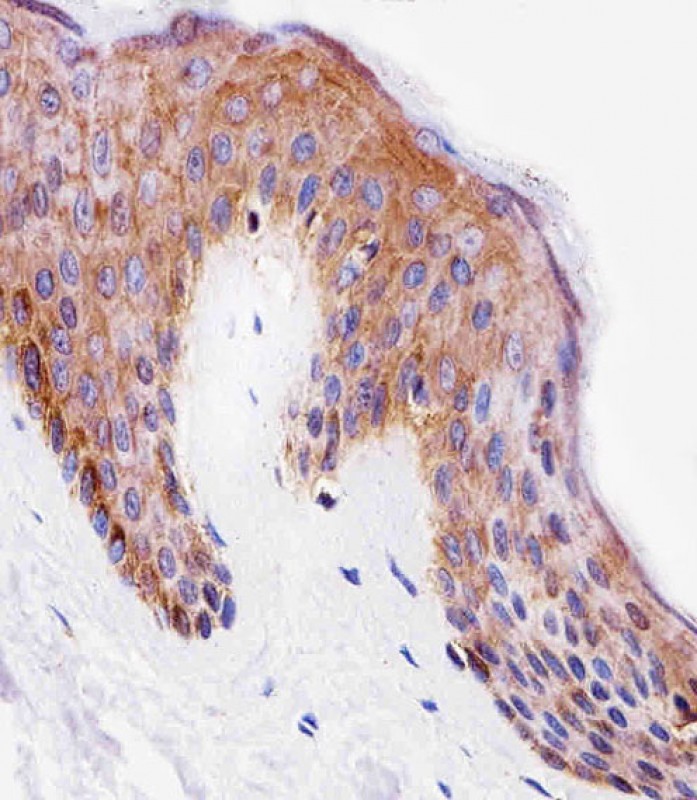
Immunohistochemical analysis of paraffin-embedded H. skin section using PITPNA Antibody (C-term). BD-PB4704 was diluted at 1:25 dilution. A undiluted biotinylated goat polyvalent antibody was used as the secondary, followed by DAB staining.
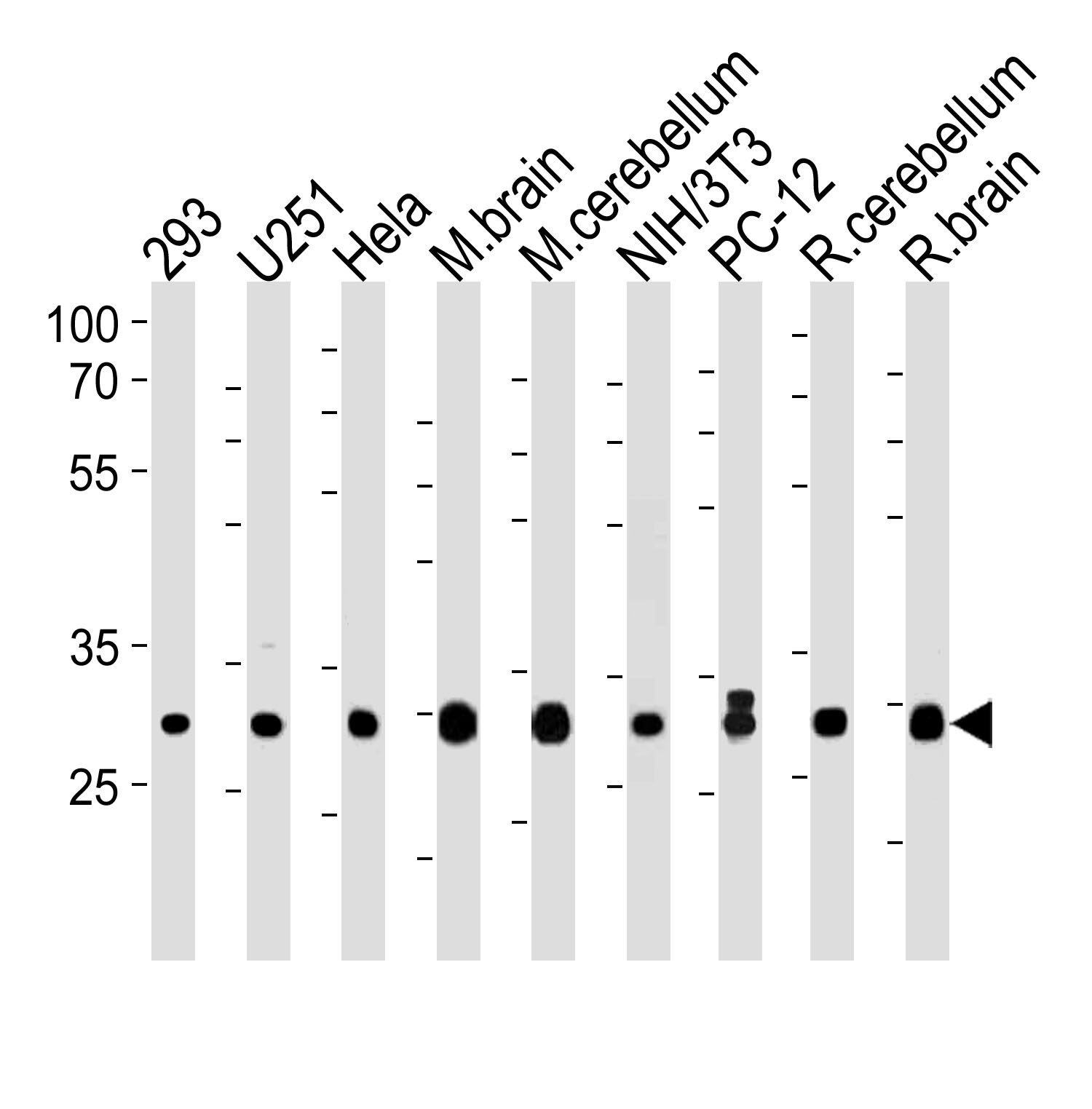
PITPNA Antibody (C-term) western blot analysis in 293,U251,Hela,mouse NIH/3T3 and rat PC12 cell line and mouse brain cerebellum,rat cerebellum and brain lysates (35ug/lane).This demonstrates the PITPNA antibody detected the PITPNA protein (arrow).
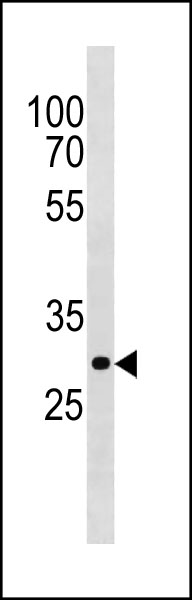
PITPNA Antibody (C-term) western blot analysis in 293 cell line lysates (35ug/lane).This demonstrates the PITPNA antibody detected the PITPNA protein (arrow).
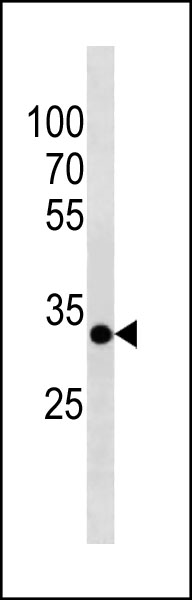
PITPNA Antibody (C-term) western blot analysis in mouse brain tissue lysates (35ug/lane).This demonstrates the PITPNA antibody detected the PITPNA protein (arrow).
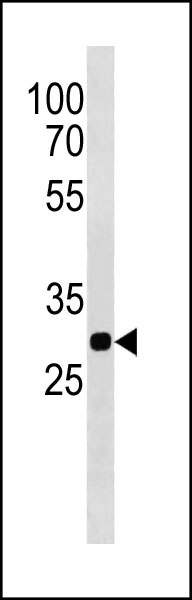
PITPNA Antibody (C-term) western blot analysis in rat cerebellum tissue lysates (35ug/lane).This demonstrates the PITPNA antibody detected the PITPNA protein (arrow).
相关文献
产品问答
相关产品

市场:027-65023363 行政/人事:027-62439686 邮箱:marketing@brainvta.com 客服:18140661572(活动咨询、售后反馈等)
销售总监:张经理 18995532642 华东区:陈经理 18013970337 华南区:王经理 13100653525 华中/西区:杨经理 18186518905 华北区:张经理 18893721749
地址:中国武汉东湖高新区光谷七路128号中科开物产业园1号楼
Copyright © 武汉枢密脑科学技术有限公司. All RIGHTS RESERVED.
鄂ICP备2021009124号 DIGITAL BY VTHINK